
Daniil Litvinov
Bioinformatician | Data scientist

Bioinformatician | Data scientist
I am a computational biologist with a commercial experience in python programming and in machine learning.
My industry experience is applying machine learning methods and statistics for analyzing omics data and computer vision methods for cryo-EM data analysis. In addition, I have experience teaching statistics and machine learning for students of biology and medicine.
My primary goal is to continue to develop my skills in the field of AI/ML/DL applied to biology.
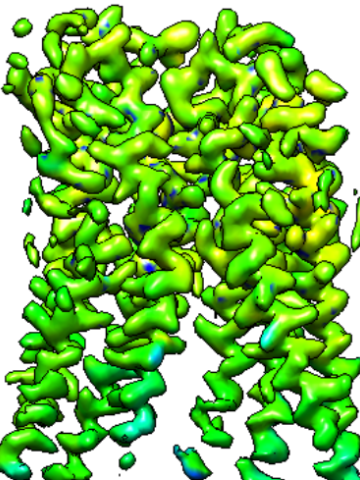
September 2022
Created CLI tool as well as a web application for cryo-EM maps resolution estimation using deep learning.
This tool is based on 3D-UNet model architecture. Classic UNet is commonly used for image segmentation tasks and it is essentially a classification, but in this case, I used this model for regression.

October 2022
This is a Python Django-based personal portfolio website.
The website uses Wagtail CMS. Wagtail is a Django Content Management System.
All content: personal information, portfolio projects, social media links, etc. can be adjusted in Wagtail admin.
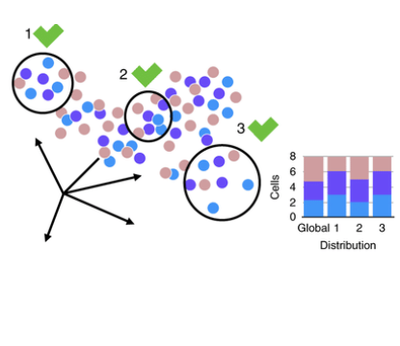
June 2021
The goal of this project is to tackle the complexity of data analysis by identifying the best approaches. The single-cell transcriptomics analysis has multiple steps, but we have focused on data integration — a crucial step when working with clinical data coming from patients.

February 2021
This project aims to study differential genes expression of 19 sportsmen during physical and psychological stress before and after running in extreme highlands conditions (2450-3450 m, Elbrus m.) and also in the "start" point before arrival at the competition (St. Petersburg).

October 2022
Content-based recommender system API based on the text of the post and user data.
Developed models based on text features obtained with TF-IDF, BERT, RoBERTa, and DistilBERT.
Created an A/B testing system to compare models using the hit rate metric.
Python (Numpy, Pandas, Matplotlib, Seaborn, Sklearn, PyTorch, Keras, FastAPI, Django)
R (ggplot2, Seurat, DeSeq2, dplyr)
Linux, Bash, git, Docker, Kubernetes
JavaScript
Classical Machine Learning (linear models, tree-based approaches, Catboost, LightGBM, XGBoost, Bayesian methods)
Deep learning (MLP, CNN, image segmentation, detection, RNN, LSTM, Transformers, AE, VAE, GAN, TabNet)
Model tuning (Optuna, genetic algorithm, Boruta)
Interpretable machine learning (SHAP, LIME, Pixel Attribution)
Databases (NCBI, UniProt, PDB, MsigDB, SILVA)
Command-line tools (Cellranger, cellSNP, Picard, BLAST, GATK, STAR, SPAdes)
Protein sequence analysis tools (MAFFT, MUSCLE, HMMER, ESM)
Protein structure analysis tools (Rosetta, Phenix, Coot, AlphaFold)
Russian – Native
English – Full professional proficiency
German – Elementary proficiency
Biological education, which helps me to understand specialized biological and medical literature
Work experience in molecular, microbiological, and biochemical labs
Agile software development methods
Presentation skills
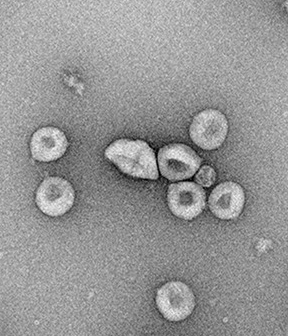
Structural characterization of propiolactone inactivated severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) particles
2022
Here, we characterized the β-propiolactone inactivated SARS-CoV-2 virions using transmission electron microscopy (TEM) and atomic force microscopy (AFM).
TEM showed that the spike (S) proteins were in the pre-fusion conformation. Notably, the S proteins could be recognized by specific monoclonal antibodies. Analytical TEM showed that the inactivated virions retained nucleic acid. Altogether, we demonstrated that the inactivated SARS-CoV-2 virions retain the structural features of native viruses and provide a prospective vaccine candidate.
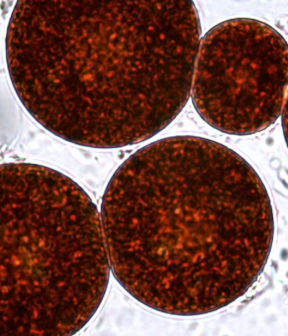
Combined Production of Astaxanthin and Carotene in a New Strain of the Microalga Bracteacoccus aggregatus BM5/15 (IPPAS C-2045)
2021
Possible pathways of astaxanthin synthesis are proposed based on the carotenoid composition obtained in this work.
Collectively, a new strain B. aggregatus BM5/15 is a potential biotechnological source of two natural antioxidants, astaxanthin and β-carotene. The results give rise for further works on the optimization of B. aggregatus cultivation on an industrial scale.
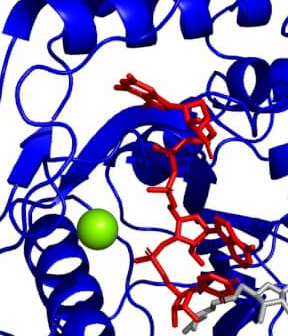
Molecular Modeling of the HR2 and Transmembrane Domains of the SARS-CoV-2 S Protein in the Prefusion State (2021)
2021
The paper reports molecular modeling of the S protein fragment corresponding to its coiled-coil HR2 domain and fully palmitoylated transmembrane domain.
Model stability in the lipid bilayer was confirmed by all-atom and coarse-grained molecular dynamics simulations. It has been demonstrated that palmitoylation leads to a significant decrease in transmembrane domain mobility and local bilayer thickening, which may be relevant for protein trimerization.
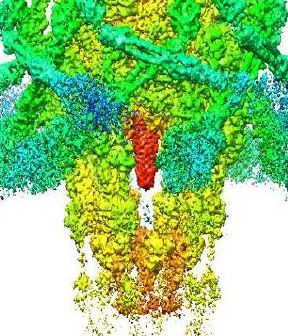
Structure of A. Baumannii Phage Tapaz, Revealed with Cryo-Electron Microscopy
2021
The cryo-EM map was reconstructed with single particle analysis independently for the capsid, tail, and baseplate regions.
The capsid was reconstructed at 3.9 Å resolution with I3 symmetry applied. The baseplate region of the phage was reconstructed at 3.5 Å resolution with C3 symmetry. The tail region was reconstructed at 2.6 Å resolution with helical symmetry (Rise 36.4 Å, Twist 25.7 deg).
The initial atomic model for the tail region was built from the sequence with Deeptracer and was further refined in coot.

Diversity of Carotenogenic Microalgae in the White Sea Polar Region
2020
We isolated several strains of carotenogenic microalgae from the coastal zone of the White Sea, where they were abundant.
The obtained microalgae related to four species of Chlorophytes: Haematococcus lacustris, H. rubicundus, Coelastrella aeroterrestrica, and Bracteacoccus aggregatus. The last three species have been reported for polar latitudes for the first time.
Most likely, carotenogenic algae on the White Sea coast are abundant due to their high physiological and metabolic plasticity, which is essential for surviving under adverse conditions of the northern regions.
© 2022. Designed and developed by Daniil Litvinov